nn.py¶
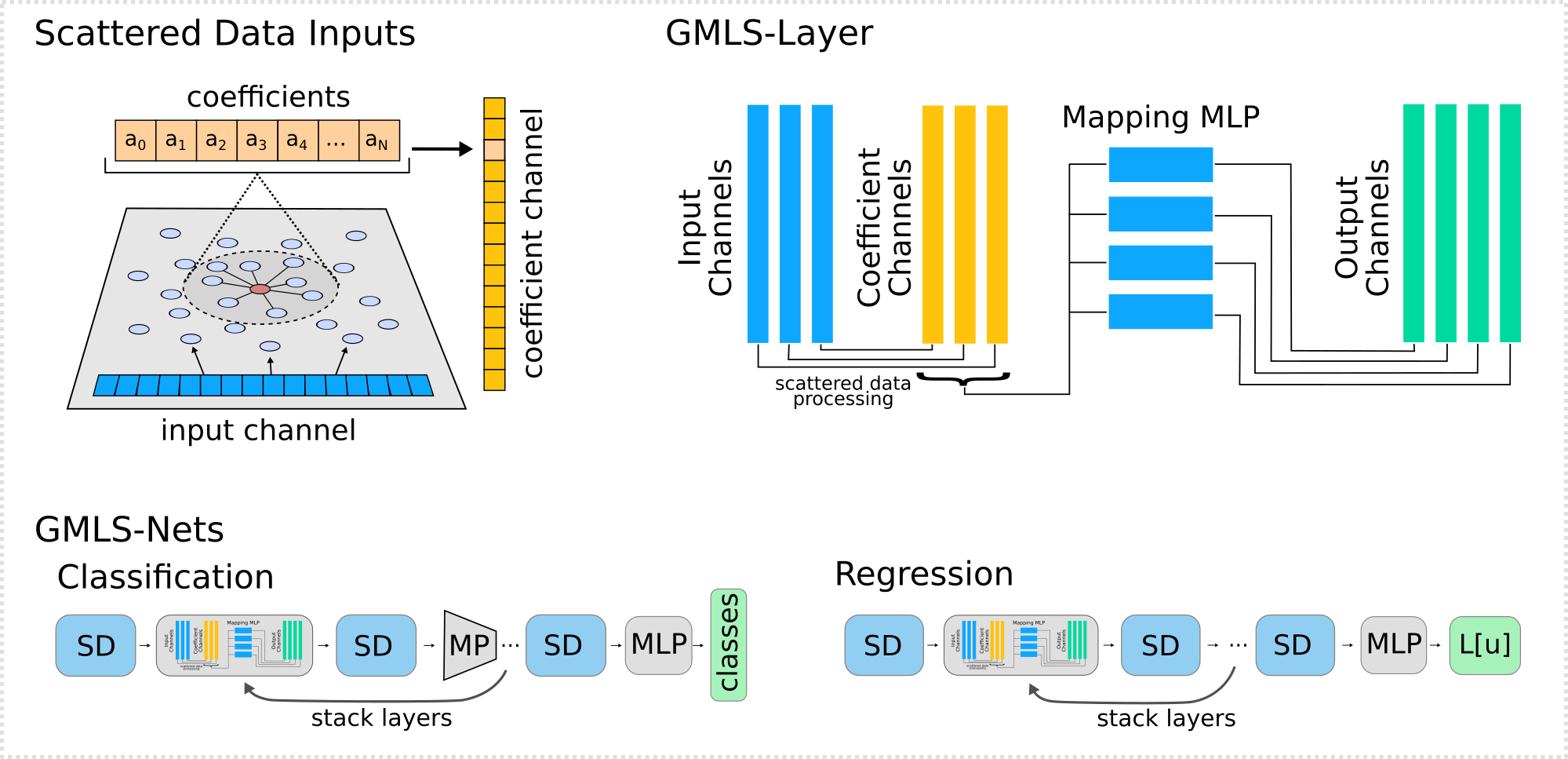
PyTorch implementation of GMLS-Nets. Module for neural networks for processing scattered data sets using Generalized Moving Least Squares (GMLS).
If you find these codes or methods helpful for your project, please cite:
-
class
nn.ExtractFromTuple(index=0)¶ Extracts from a tuple of outputs one of the components.
-
__init__(index=0)¶ Initializes the index to extract.
-
__module__= 'nn'¶
-
forward(input)¶ Extracts the tuple entry with the specified index.
-
-
class
nn.ExtractFromTuple_Function¶ Extracts from a tuple of outputs one of the components.
-
__module__= 'nn'¶
-
static
backward(ctx, grad_output)¶ Computes gradient of the extraction.
-
static
forward(ctx, input, index)¶ Extracts tuple entry with the specified index.
-
-
class
nn.GMLS_Layer(flag_case, porder, pts_x1, epsilon, weight_func, weight_func_params, mlp_q=None, pts_x2=None, device=None, flag_verbose=0)¶ The GMLS-Layer processes scattered data by using Generalized Moving Least Squares (GMLS) to construct a local reconstruction of the data (here polynomials). This is represented by coefficients that are mapped to approximate the action of linear or non-linear operators on the input field.
As depicted above, the architecture processes a collection of input channels into intermediate coefficient channels. The coefficient channels are then collectively mapped to output channels. The mappings can be any unit for which back-propagation can be performed. This includes linear layers or non-linear maps based on multilayer perceptrons (MLPs).
Examples
Here is a typical way to construct a GMLS-Layer. This is done in the following stages.
(i)Construct the scattered data locations xj, xi at which processing will occur. Here, we create points in 2D.>>> xj = torch.randn((100,2),device=device); xi = torch.randn((100,2),device=device);
(ii)Construct the mapping unit that will be applied pointwise. Here we create an MLP with Nc input coefficient channels and channels_out output channels.>>> layer_sizes = []; >>> num_input = Nc*num_polys; # number of channels (NC) X number polynomials (num_polys) (cross-channel coupling allowed) >>> num_depth = 4; num_hidden = 100; channels_out = 16; # depth, width, number of output filters >>> layer_sizes.append(num_polys); >>> for k in range(num_depth): >>> layer_sizes.append(num_hidden); >>> layer_sizes.append(1); # a single unit always gives scalar output, we then use channels_out units. >>> mlp_q_map1 = gmlsnets_pytorch.nn.MLP_Pointwise(layer_sizes,channels_out=channels_out);
(iii)Create the GMLS-Layer using these components.>>> weight_func1 = gmlsnets_pytorch.nn.MapToPoly_Function.weight_one_minus_r; >>> weight_func_params = {'epsilon':1e-3,'p'=4}; >>> gmls_layer_params = { 'flag_case':'standard','porder':4,'Nc':3, 'mlp_q1':mlp_q_map1, 'pts_x1':xj,'pts_x2':xi,'epsilon':1e-3, 'weight_func1':weight_func1,'weight_func1_params':weight_func1_params, 'device':device,'flag_verbose':0 }; >>> gmls_layer=gmlsnets_pytorch.nn.GMLS_Layer(**gmls_layer_params);
Here is an example of how a GMLS-Layer and other modules in this package can be used to process scattered data. This could be part of a larger neural network in practice (see example codes for more information). For instance,
>>> layer1 = nn.Sequential(gmls_layer, # produces output tuple of tensors (ci,xi) with shapes ([batch,ci,xi],[xi]). #PdbSetTraceLayer(), ExtractFromTuple(index=0), # from output keep only the ui part and discard the xi part. #PdbSetTraceLayer(), PermuteLayer((0,2,1)) # organize indexing to be [batch,xi,ci], for further processing. ).to(device);
You can uncomment the PdbSetTraceLayer() to get breakpoints for state information and tensor shapes during processing. The PermuteLayer() changes the order of the indexing. Also can use ReshapeLayer() to reshape the tensors, which is especially useful for processing data related to CNNs.
Much of the construction can be further simplified by writing a few wrapper classes for your most common use cases.
More information also can be found in the example codes directory.
-
__init__(flag_case, porder, pts_x1, epsilon, weight_func, weight_func_params, mlp_q=None, pts_x2=None, device=None, flag_verbose=0)¶ Initializes the GMLS layer.
- Parameters
flag_case (str) – Flag for the type of architecture to use (default is ‘standard’).
porder (int) – Order of the basis to use (polynomial degree).
pts_x1 (Tensor) – The collection of domain points \(x_j\).
epsilon (float) – The \(\epsilon\)-neighborhood size to use to sort points (should be compatible with choice of weight_func_params).
weight_func (func) – Weight function to use.
weight_func_params (dict) – Weight function parameters.
mlp_q (module) – Mapping q unit for computing \(q(c)\), where c are the coefficients.
pts_x2 (Tensor) – The collection of target points \(x_i\).
device – Device on which to perform calculations (GPU or other, default is CPU).
flag_verbose (int) – Level of reporting on progress during the calculations.
-
__module__= 'nn'¶
-
forward(input)¶ Computes GMLS-Layer processing scattered data input field uj to obtain output field vi.
- Parameters
input (Tensor) – Input channels uj organized in the shape [batch,xj,uj].
- Returns
The output channels and point locations (vi,xi). The field vi = q(ci).
- Return type
tuple
-
to(device)¶ Moves data to GPU or other specified device.
-
-
class
nn.MLP1(layer_sizes, flag_bias=True, flag_verbose=0)¶ Creates a multilayer perceptron (MLP).
-
__init__(layer_sizes, flag_bias=True, flag_verbose=0)¶ Initializes MLP and specified layer sizes.
-
__module__= 'nn'¶
-
forward(input, params=None)¶ Applies the MLP to the input data.
- Parameters
input (Tensor) – The coefficient channel data organized as one stacked
of size Nc*M, where Nc is number of channels and M is number of (vector) –
per channel. (coefficients) –
- Returns
The evaluation of the network. Returns tensor of size [batch,1].
- Return type
Tensor
-
to(device)¶ Moves data to GPU or other specified device.
-
-
class
nn.MLP_Pointwise(layer_sizes, channels_in=1, channels_out=1, flag_bias=True, flag_verbose=0)¶ Creates a collection of multilayer perceptrons (MLPs) for each output channel. The MLPs are then applied at each target point xi.
-
__init__(layer_sizes, channels_in=1, channels_out=1, flag_bias=True, flag_verbose=0)¶ Initializes the structure of the pointwise MLP module with layer sizes, number input channels, number of output channels.
- Parameters
layer_sizes (list) – The number of hidden units in each layer.
channels_in (int) – The number of input channels.
channels_out (int) – The number of output channels.
flag_bias (bool) – If the MLP should include the additive bias b added into layers.
flag_verbose (int) – The level of messages generated on progress of the calculation.
-
__module__= 'nn'¶
-
create_mlp_unit(layer_sizes, unit_name='', flag_bias=True)¶ Creates an instance of an MLP with specified layer sizes.
-
forward(input, params=None)¶ Applies the specified MLP pointwise to the collection of input data to produce pointwise entries of the output channels.
-
to(device)¶ Moves data to GPU or other specified device.
-
-
class
nn.MapToPoly(porder, weight_func, weight_func_params, pts_x1, epsilon=None, pts_x2=None, tree_points=None, device=None, flag_verbose=0, **extra_params)¶ This layer processes a collection of scattered data points consisting of a collection of values \(u_j\) at points \(x_j\). For a collection of target points \(x_i\), local least-squares problems are solved for obtaining a local representation of the data over a polynomial space. The layer outputs a collection of polynomial coefficients \(c(x_i)\) at each point and the collection of target points \(x_i\).
-
__init__(porder, weight_func, weight_func_params, pts_x1, epsilon=None, pts_x2=None, tree_points=None, device=None, flag_verbose=0, **extra_params)¶ Initializes the layer for mapping between field data uj at points xj to the local polynomial reconstruction represented by coefficients ci at target points xi.
- Parameters
porder (int) – Order of the basis to use. For polynomial basis is the degree.
weight_func (func) – Weight function to use.
weight_func_params (dict) – Weight function parameters.
pts_x1 (Tensor) – The collection of domain points \(x_j\).
epsilon (float) – The \(\epsilon\)-neighborhood size to use to sort points (should be compatible with choice of weight_func_params).
pts_x2 (Tensor) – The collection of target points \(x_i\).
tree_points (dict) – Stored data to help speed up repeated calculations.
device – Device on which to perform calculations (GPU or other, default is CPU).
flag_verbose (int) – Level of reporting on progress during the calculations.
**extra_params – Extra parameters allowing for specifying layer name and caching mode.
-
__module__= 'nn'¶
-
eval_poly(pts_x, pts_x2_i0, c_star_i0, porder=None, flag_verbose=None)¶ Evaluates the polynomial reconstruction around a given target point pts_x2_i0.
-
extra_repr()¶ Displays information associated with this module.
-
forward(input)¶ For a field u specified at points xj, performs the mapping to coefficients c at points xi, (uj,xj) \(\rightarrow\) (ci,xi).
-
load_cache_data(cache_filename)¶ Load the needed matrices and related data from .pickle. (Warning: prototype codes here currently and not tested).
-
save_cache_data(cache_filename)¶ Save needed matrices and related data to .pickle for later cached use. (Warning: prototype codes here currently and not tested).
-
to(device)¶ Moves data to GPU or other specified device.
-
-
class
nn.MapToPoly_Function¶ This layer processes a collection of scattered data points consisting of a collection of values \(u_j\) at points \(x_j\). For a collection of target points \(x_i\), local least-squares problems are solved for obtaining a local representation of the data over a polynomial space. The layer outputs a collection of polynomial coefficients \(c(x_i)\) at each point and the collection of target points \(x_i\).
-
__module__= 'nn'¶
-
static
backward(ctx, grad_output, grad_pts_x2)¶ Consider a field u specified at points xj and the mapping to coefficients c at points xi, (uj,xj) –> (ci,xi). Computes the gradient of the mapping for backward propagation.
-
static
eval_poly(pts_x, pts_x2_i0, c_star_i0, porder, flag_verbose)¶ Evaluates the polynomials locally around a target point xi given coefficients c.
-
static
forward(ctx, input, porder, weight_func, weight_func_params, pts_x1, epsilon=None, pts_x2=None, cached_data=None, tree_points=None, device=None, flag_verbose=0)¶ For a field u specified at points xj, performs the mapping to coefficients c at points xi, (uj,xj) \(\rightarrow\) (ci,xi).
- Parameters
input (Tensor) – The input field data uj.
porder (int) – Order of the basis to use (polynomial degree).
weight_func (function) – Weight function to use.
weight_func_params (dict) – Weight function parameters.
pts_x1 (Tensor) – The collection of domain points \(x_j\).
epsilon (float) – The \(\epsilon\)-neighborhood size to use to sort points (should be compatible with choice of weight_func_params).
pts_x2 (Tensor) – The collection of target points \(x_i\).
cache_data (dict) – Stored data to help speed up repeated calculations.
tree_points (dict) – Stored data to help speed up repeated calculations.
device (torch.device) – Device on which to perform calculations (GPU or other, default is CPU).
flag_verbose (int) – Level of reporting on progress during the calculations.
- Returns
The coefficient values ci at the target points xi. The target points xi.
- Return type
tuple of (ci,xi)
-
static
generate_mapping(weight_func, weight_func_params, porder, epsilon, pts_x1, pts_x2, tree_points=None, device=None, flag_verbose=0)¶ Generates for caching the data for the mapping from field values (uj,xj) \(\rightarrow\) (ci,xi). This help optimize codes and speed up later calculations that are done repeatedly.
-
static
get_num_polys(porder, num_dim=None)¶ Returns the number of polynomials of given porder.
-
static
get_poly_1D_u(u, porder, weight_func, weight_func_params, pts_x1, epsilon=None, pts_x2=None, cached_data=None, tree_points=None, device=None, flag_verbose=0)¶ Compute the polynomial coefficients in the case of a scalar field. Would not typically call directly, used for internal purposes.
-
static
weight_one_minus_r(z1, z2, params)¶ Weight function \(\omega(x_j,x_i) = \left(1 - r/\epsilon\right)^{\bar{p}}_+.\)
- Parameters
z1 (Tensor) – The first point. Tensor of shape [1,num_dims].
z2 (Tensor) – The second point. Tensor of shape [1,num_dims].
params (dict) – The parameters are ‘p’ for decay power and ‘epsilon’ for support size.
- Returns
The weight evaluation over points.
- Return type
Tensor
-
-
class
nn.MaxPoolOverPoints(pts_x1, epsilon=None, pts_x2=None, indices_xj_i_cache=None, tree_points=None, device=None, flag_verbose=0, **extra_params)¶ Applies a max-pooling operation to obtain values \(v_i = \max_{j \in \mathcal{N}_i(\epsilon)} \{u_j\}.\)
-
__init__(pts_x1, epsilon=None, pts_x2=None, indices_xj_i_cache=None, tree_points=None, device=None, flag_verbose=0, **extra_params)¶ Setup of max-pooling operation.
- Parameters
pts_x1 (Tensor) – The collection of domain points \(x_j\). We assume size [num_pts,num_dim].
epsilon (float) – The \(\epsilon\)-neighborhood size to use to sort points (should be compatible with choice of weight_func_params).
pts_x2 (Tensor) – The collection of target points \(x_i\).
indices_xj_i_cache (dict) – Stored data to help speed up repeated calculations.
tree_points (dict) – Stored data to help speed up repeated calculations.
device – Device on which to perform calculations (GPU or other, default is CPU).
flag_verbose (int) – Level of reporting on progress during the calculations.
**extra_params (dict) – Extra parameters allowing for specifying layer name and caching mode.
-
__module__= 'nn'¶
-
extra_repr()¶ Displays information associated with this module.
-
forward(input)¶ Applies a max-pooling operation to obtain values \(v_i = \max_{j \in \mathcal{N}_i(\epsilon)} \{u_j\}.\)
- Parameters
input (Tensor) – The collection uj of field values at the points xj.
- Returns
The collection of field values vi at the target points xi.
- Return type
Tensor
-
load_cache_data(cache_filename)¶ Load data to .pickle file for caching. (Warning: Prototype placeholder code.)
-
save_cache_data(cache_filename)¶ Save data to .pickle file for caching. (Warning: Prototype placeholder code.)
-
to(device)¶ Moves data to GPU or other specified device.
-
-
class
nn.MaxPoolOverPoints_Function¶ Applies a max-pooling operation to obtain values \(v_i = \max_{j \in \mathcal{N}_i(\epsilon)} \{u_j\}.\)
-
__module__= 'nn'¶
-
static
backward(ctx, grad_output, grad_pts_x2)¶ Compute gradients of the max pool operations from values at points (uj,xj) –> (max_ui,xi).
-
static
forward(ctx, input, pts_x1, epsilon=None, pts_x2=None, indices_xj_i_cache=None, tree_points=None, flag_verbose=0)¶ Compute max pool operation from values at points (uj,xj) to obtain (vi,xi).
- Parameters
input (Tensor) – The uj values at the location of points xj.
pts_x1 (Tensor) – The collection of domain points \(x_j\).
epsilon (float) – The \(\epsilon\)-neighborhood size to use to sort points (should be compatible with choice of weight_func_params).
pts_x2 (Tensor) – The collection of target points \(x_i\).
tree_points (dict) – Stored data to help speed up repeated calculations.
flag_verbose (int) – Level of reporting on progress during the calculations.
- Returns
The collection ui at target points (same size as uj in the non-j indices). The collection xi of target points. Tuple of form (ui,xi).
- Return type
tuple
Note
We assume that all inputs are pytorch tensors with pts_x1.shape = [num_pts,num_dim] and similarly for pts_x2.
-
-
class
nn.PdbSetTraceLayer¶ Allows for placing break-points within the call sequence of layers using pdb.set_trace(). Helpful for debugging networks.
-
__init__()¶ Initialization (currently nothing to do, but call super-class).
-
__module__= 'nn'¶
-
forward(input)¶ Executes a PDB breakpoint inside of a running network to help with debugging.
-
-
class
nn.PermuteLayer(permute=None)¶ Performs permutation of indices of a tensor output within a network.
-
__init__(permute=None)¶ Initializes the indexing permuation to apply to tensors.
-
__module__= 'nn'¶
-
forward(input)¶ Applies and indexing permuation to the input tensor.
-
-
class
nn.ReshapeLayer(reshape, permute=None)¶ Performs reshaping of a tensor output within a network.
-
__init__(reshape, permute=None)¶ Initializes the reshaping form to use followed by the indexing permulation to apply.
-
__module__= 'nn'¶
-
forward(input)¶ Reshapes the tensor followed by applying a permutation to the indexing.
-