Polymer Example
Details:
More details and files for this example can be found on GitHub at:
GitHub: mango-selm
- Quick Start:
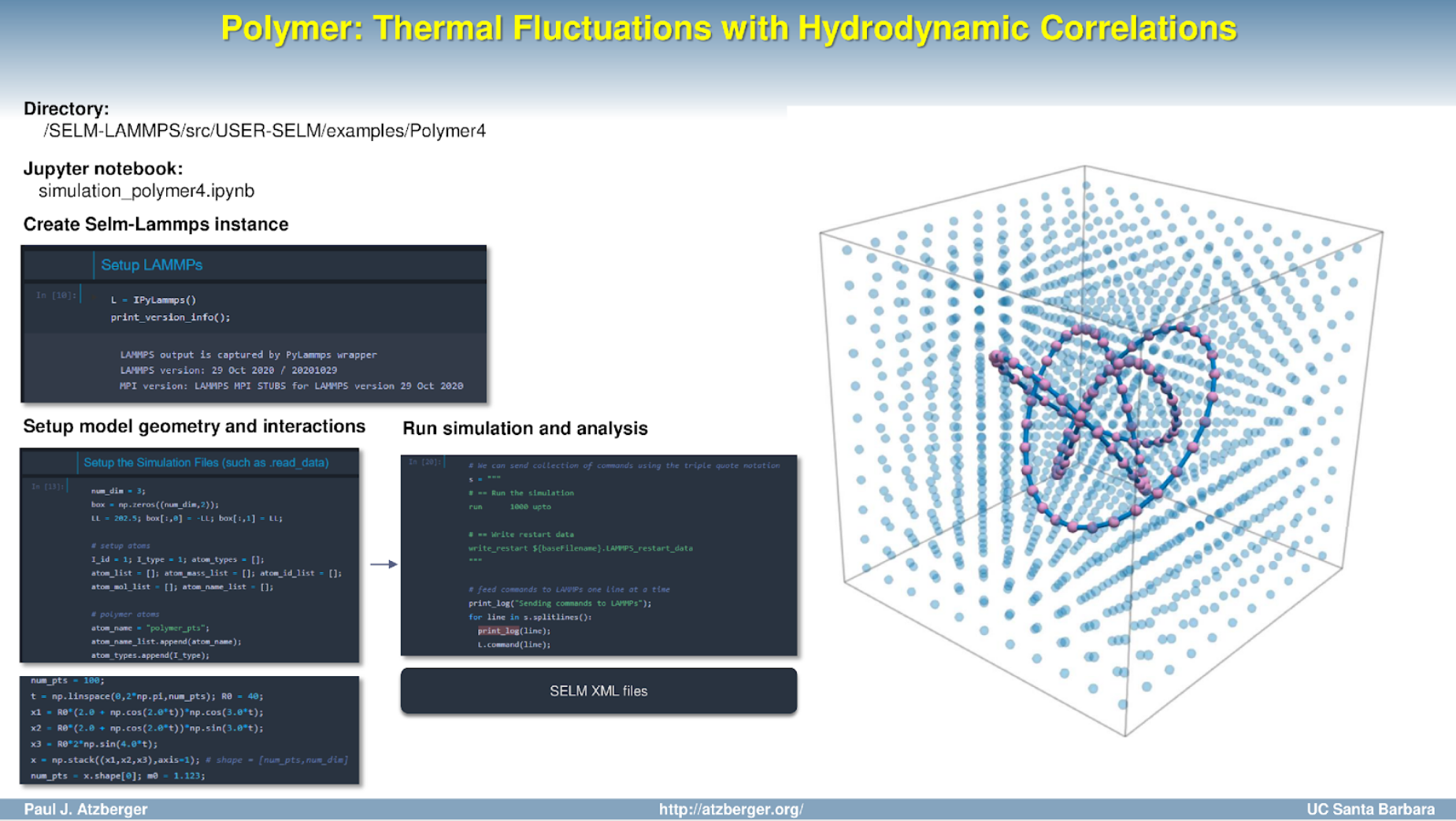
- Open Jupyter Notebook simulation_polymer4.ipynb.
- Or run Python script python simulation_polymer4.py.
- Simulation output is in the directory ./output/<sim_name>/<case_name>. Paraview can be used for visualization of the VTK (.vtp/.vtu) output files. VMD can be used for .dcd files. See the visualization scripts in the output directory for examples.
Details:
Quick start can try "pip install selm-lammps" for pre-compiled binaries for select platforms (i.e. latest version of Ubuntu).
The Jupyter Notebook and Python scripts make use of the liblammps.so shared library and lammps.py module. This should be installed in the <python_env>/lib/pythonX.X/site-packages/selm-lammps directory of your python installation (see README in /bin directory for details and pre-compiled binaries for platforms).
The Python interfaces creates a LAMMPS instance using "L = PyLammps()" which is then issued commands using "L.command(<cmd_str>)". The scripts generate the model geometry, LAMMPS "read_data" files, and issue the commands to setup and run the simulation.
The SELM package is controlled via the command cmd_str = "fix SELM paramFile.SELM_params". The file paramFile.SELM_params can be considered the "master file" playing for the SELM package a role coordinating the package behaviors. All of the SELM package settings are controlled through XML files of the form *.SELM_*. These give the settings for the Eulerian degrees of freedom (solvent fluid), the Lagrangian degrees of freedom (particles), and how they are coupled.
- In this example, XML template files that are used for each simulation are located in the /Model1/ sub-directory. You can edit XML there to adjust SELM parameters.
For further discussions of the steps for setting up this simulation, see the comments in the Jupyter Notebook or Python script. Also, see the Related Talks Section for videos and slides.
The codes can be obtained from:
