Investigations in Biology:
Experimental advances are yielding a wealth of quantitative information about the mechanics and localization of processes in cell biology. This is both at the level of single molecules and at more collective levels. This presents many opportunities for theoretical approaches to play a role in the study of biological systems. In collaboration with experimentalists on-campus at UCSB, we are working on several projects.
Motor Proteins:
Motor proteins act essentially as microscopic machines within cells to form cellular structures, transport materials, or generate forces. We are using stochastic modeling, analysis, and simulations to study the basic mechanisms underlying how molecular motor proteins may operate. In this work, we are making a particular effort toward building models for the analysis of specific experimental data sets. In previous work we have studied the kinesin motor protein and developed a coarse-grained mechanical model consistent with available optical trap experimental data, see papers.
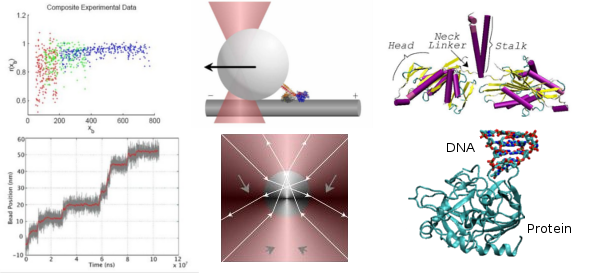
Microtubules:
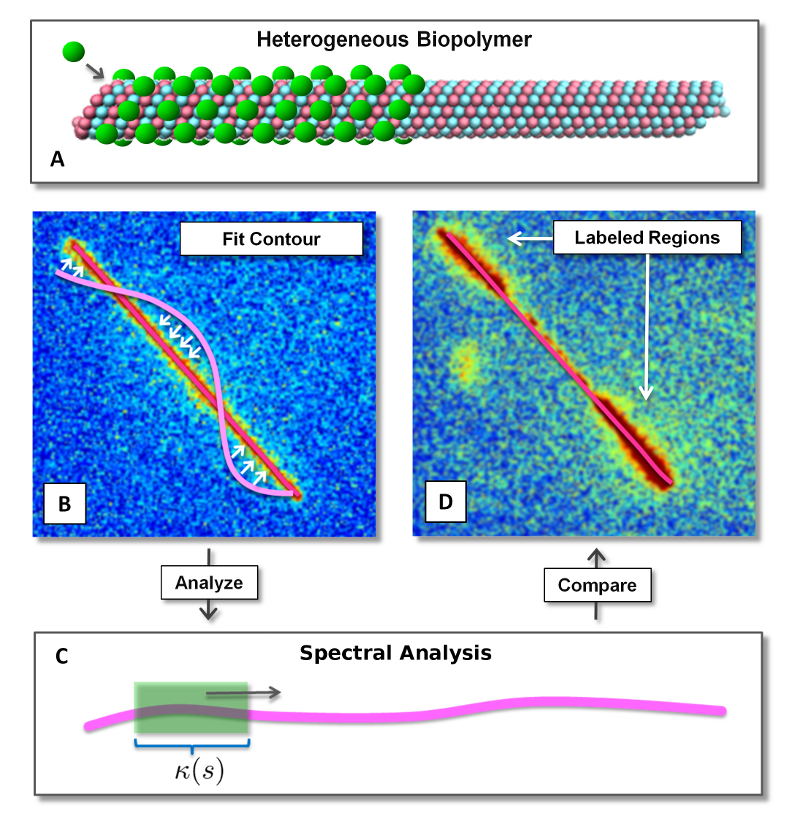
Microtubules are long polymer filaments of the cytoskeleton that play many important roles in cell biology. This includes allowing cells to resist deformation during mechanical loading, serving as a highway for many types of motor proteins, and sustaining forces responsible for the separation of chromosomes during cell division. We are studying microtubule mechanics in collaboration with the laboratory of Megan Valentine, Department of Mechanical Engineering and the graduate student David Valdman, Department of Mathematics. In particular, we are studying how MAP proteins affect microtubule mechanics. One approach is based on the thermal fluctuations of microtubules observed through fluorescence microscopy. We are developing new approaches based on spectral analysis to infer elastic properties from observed thermal fluctuations. These methods are being used to develop new experimental assays to study how particular MAP proteins augment microtubule mechanical properties and to study biological consequences.
Aptamers:
We are studying Aptamers in collaboration with the laboratory of H.T. Soh, Department of Mechanical Engineering and undergraduate Joe Rudzinski (CCS Program, UCSB). Aptamers are short segments of DNA or RNA which bind to proteins. In applications, specific aptamers (nucleic acid sequences) are sought which bind to a desired target protein with high affinity and specificity. Applications include purification methods for proteins, analytics in biosensor devices, and target validation in pharmaceutical drug development. Obtaining aptamers with strong affinity and specificity binding a given target molecule poses a significant challenge in practice. We are developing mathematical approaches to study experimental methods for the selection of DNA-Aptamers from a library of random sequences.
Select Publications
- Force Spectroscopy of Complex Biopolymers with Heterogeneous Elasticity, D. Valdman, B. Lopez, M. T. Valentine, and P.J. Atzberger, Soft Matter, The Royal Society of Chemistry, (2013). [preprint PDF] [full paper DOI]
- Spectral Analysis Methods for the Robust Measurement of the Flexural Rigidity of Biopolymers, D. Valdman, P.J. Atzberger, D. Yu, and M. T. Valentine, Bio. Phys. J., Vol. 102, Iss. 5, pg. 1144-–1153, (2012). [preprint PDF] [full paper DOI]
- Influence of Target Concentration and Background Binding on In Vitro Selection of Affinity Reagents, J. Wang, J. F. Rudzinski, Q. H. Gonga, H.T. Soh, P.J. Atzberger (accepted 2012), [preprint PDF]
- Micromagnetic Selection of Aptamers in Microfluidic Channels, Lou, X., Qian, J., Yi, X., Viel, L., Gerdon, A.E, Lagally, E.T, Atzberger, P.J., Heeger, A.J., and Soh, H.T., PNAS, Vol. 106 No. 9 pp. 2989-2994, (2009), [DOI] [PDF]
- Analysis of Selection Approaches for Aptamer Molecular Libraries, Rudzinski, J., Soh, T., Atzberger, P.J., (technical report) (2009), [PDF]
- Stochastic Reduction Method for Biological Chemical Kinetics using Time-Scale Separation, Pahlajani, C.D., Atzberger, P.J., Khammash, M. (2010), (preprint). [PDF]
- A Brownian Dynamics Model of Kinesin in Three Dimensions Incorporating the Force-Extension Profile of the Coiled-Coil Cargo Tether, Atzberger, P.J. and Peskin, C.S., Bull. Math. Biol., vol. 68, no. 1, pp. 131-160, (2006). [PDF] [DOI]

